
Dr. Jans Aasman, CEO, Franz Inc.
LinkedIn: Jans Aasman
Dr. Richard Wallace, AI Scientist
LinkedIn: Franz Inc.

Imagine querying a medical database to analyze patients with nervous system disorders, only to have the results include unrelated conditions like heart failure and burn injuries. These errors aren’t just technical nuisances—they can have life-or-death consequences in clinical settings.
These errors can stem from the Unified Medical Language System (UMLS), a cornerstone of healthcare data interoperability that unifies over 200 biomedical vocabularies, including SNOMED CT and ICD-10. Despite its critical role, the complexity of UMLS can obscure hidden cycles, potentially leading to inconsistent and inaccurate clinical conclusions.
But by combining the unique strengths of machine learning, logic-based reasoning, and Large Language Models (LLMs), we can clean up UMLS and ensure its massive knowledge graph is accurate, consistent, and usable.
The Importance of UMLS in Biomedical Informatics
UMLS plays a critical role in biomedical informatics by unifying terminologies and mapping relationships across multiple vocabularies, ensuring semantic interoperability. Its Metathesaurus integrates data from various sources, allowing healthcare systems to identify synonymous terms across different medical databases, while the Semantic Network establishes structured relationships between concepts for efficient information retrieval.
Healthcare providers, researchers, and developers rely on UMLS to standardize medical concepts, improve patient care coordination, and enable advanced research. However, inconsistencies in UMLS can lead to incorrect or inefficient results, particularly in systems using symbolic reasoning and hierarchical queries.
The Challenge of Cycles in UMLS
A key issue with the UMLS is the presence of cycles in its relationship structures, particularly in its “narrower” relationships that form the foundation of many hierarchical queries. A cycle occurs when a concept in the UMLS graph is related to itself through a series of narrower or broader relationships. For example, a query designed to retrieve all more specific diagnoses under “nervous system disorder” might mistakenly include unrelated concepts like “heart failure” or “renal dysplasia” due to cycles in the graph.
These cycles not only lead to erroneous query results but also impede the use of UMLS in automated deduction and reasoning, limiting its effectiveness in real-world applications. As biomedical informatics increasingly relies on advanced data analytics, these cycles can cause combinatorial explosions in query complexity, further reducing the system’s efficiency.
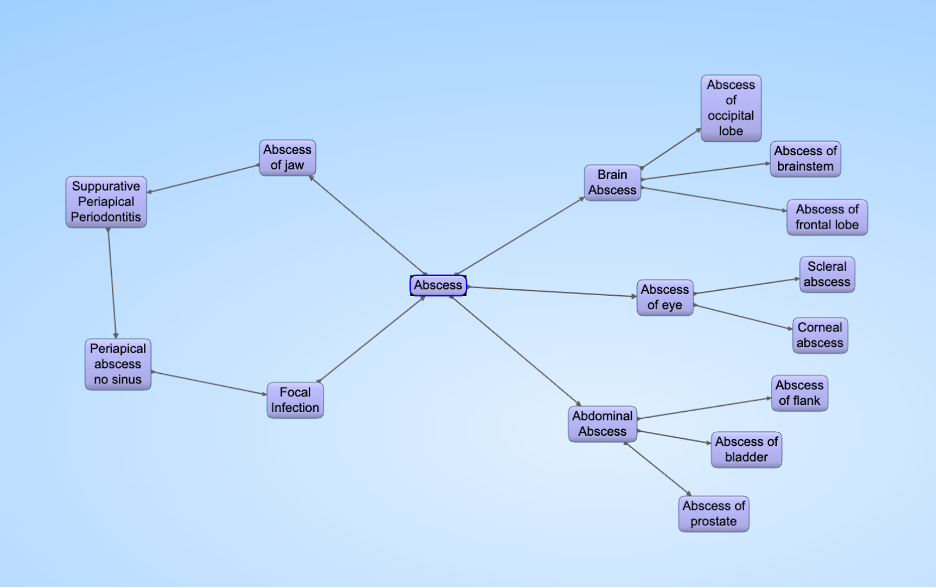
A disease cycle in UMLS. When a query asks for all subsets of “Abscess of Jaw”, the query cycles through “Abscess” and will include conditions like “Abscess of prostate.”
A Neuro-Symbolic Approach to Cleaning UMLS
By applying a Neuro-Symbolic AI approach that integrates machine learning models, symbolic reasoning, and LLMs, we can systematically identify and eliminate cycles within the UMLS graph, ensuring its integrity.
Neuro-Symbolic AI leverages LLMs’ ability to process text-based information. By generating queries that ask whether one concept is truly a narrower subset of another (e.g., “Is congestive heart failure a subset of heart diseases?”), the system can validate relationships and identify incorrect or inconsistent connections that form cycles. When a cycle is detected, the system can prune the invalid relationships, effectively breaking the cycle and restoring the graph to a valid, directed acyclic structure.
This Neuro-Symbolic approach offers significant advantages over previous methods, which often relied on labor-intensive manual audits or simpler rule-based systems that could not fully account for the complexity of UMLS. By combining symbolic reasoning (to handle structured data) with machine learning (to adapt to new information and handle unstructured data), this hybrid approach offers a more powerful and scalable solution.
Methodology for Cycle Detection and Elimination
The process of identifying and eliminating cycles in the UMLS knowledge graph involves several key steps:
- Convert UMLS to RDF Triples: The UMLS is first transformed into a graph-based structure using Resource Description Framework (RDF) triples, making it easier to query and analyze the relationships between concepts.
- Explore for Cycles: Using a depth-first search algorithm, the system examines the UMLS graph for cycles, specifically focusing on transitive relationships like “narrower” and “broader.”
- LLM-Based Validation: Once a cycle is detected, LLMs are prompted with questions such as “Does concept X describe a narrower subset of concept Y?” The LLM’s answer (“yes” or “no”) is used to validate the relationship. If the answer suggests that the relationship is invalid, that edge is marked for deletion.
- Cycle Pruning: After marking invalid relationships, they are deleted from the graph, and the process is repeated for more complex cycles involving more than two nodes.
- Re-running the Procedure: Once cycles are removed, the system continuously re-evaluates the graph to ensure no further cycles are introduced and that the remaining relationships are valid.
By the end of this process, the Neuro-Symbolic AI framework can successfully prune nearly 70% of cycles from the UMLS, significantly improving its reliability for hierarchical queries and symbolic reasoning applications.
Impact of a Cleaned UMLS on Biomedical Informatics
For data scientists and healthcare professionals, a cycle-free UMLS means more accurate and reliable results from queries, improving the quality of clinical decision support systems, patient outcome predictions, and other AI-driven healthcare applications. Moreover, eliminating cycles reduces query complexity, making biomedical systems more efficient and scalable.
More importantly, by improving the precision of data analytics underlying medical treatments, a cleaned-up UMLS could potentially save lives. Inaccurate or misleading medical information can lead to poor clinical decisions, but with a more trustworthy and accurate system, healthcare providers can make better-informed choices, ultimately leading to better patient outcomes.
